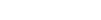
Scientists at Johns Hopkins Kimmel Cancer Center in the US have been using a computer algorithm called the Coordinate Gene Activity in Pattern Sets algorithm (CoGAPS) to study the evolution of treatment resistance in head and neck cancer.
The scientists aimed to develop new methods to examine how cancers acquire resistance to treatment and whether those changes could be modelled computationally to determine patient-specific timelines of resistance.

Discover B2B Marketing That Performs
Combine business intelligence and editorial excellence to reach engaged professionals across 36 leading media platforms.
The algorithm has been used to determine the molecular changes associated with resistance during the course of the development of the resistance. To facilitate this, the researchers developed new methods of collecting data from in vitro cell models and took a computational analysis approach to measuring observations.
Senior author Dr Elana Fertig said: “The biggest novelty in this paper is considering time as a variable. We have to prove that it matters before putting that burden on patients but we think it will result in better treatment.”
The research has been published by Genome Medicine.
The study examined cetuximab treatment effects on cancer cells from head and neck squamous cell carcinoma over an 11-week period. The same pool of cells was used to see what happened during the time period in an attempt to avoid any outside variables from using different batches of cells.
US Tariffs are shifting - will you react or anticipate?
Don’t let policy changes catch you off guard. Stay proactive with real-time data and expert analysis.
By GlobalDataCoGAPS was used to quantify evolving changes during treatment. The resulting data showed how the changes occurred over time and when the changes resulted in immediate therapeutic response or resistance. This information could lead to combined or alternative therapies to help tackle resistance.
Co-lead author Dr Genevieve Stein-O’Brien said: “Most model systems were developed to sync to existing data, comparing pre- and post-treatment. To take this algorithm and find out how the resistance was acquired, we needed to know what was going on in between the pre- and post-treatment during the full time course.”
As the CoGAPS algorithm combines experimental biology and computer programming, the researchers hope it will give doctors and patients a better indication about how the disease is progressing during treatment.
Co-lead author Dr Luciane Kagohara said the CoGAPS algorithm takes a step away from standard approaches but allows the deeper study of therapeutic resistance and individual fundamental pathways.
“If we can map that, it will really pave the way to predict when resistance is going to occur and what drugs can be used to combat that resistance,” she added.
The scientists hope their computational approach will be used in the future for other types of cancers and other drug therapies.