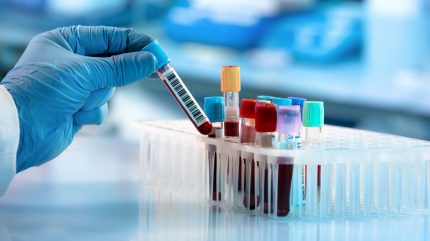
Researchers from Weill Cornell Medicine (WCM) and New York Genome Center (NYGC), US, have identified an “accurate” and “sensitive” method for identifying cancer through blood tests, leveraging whole-genome sequencing and an error-correcting technique.
Through a study, the researchers assessed the performance of a new sequencing platform from Ultima Genomics, demonstrating that the platform’s affordability does not compromise the depth of coverage.

Discover B2B Marketing That Performs
Combine business intelligence and editorial excellence to reach engaged professionals across 36 leading media platforms.
This enables the detection of minute concentrations of circulating tumour DNA. The researchers further enhanced the method’s precision by incorporating an error correction mechanism.
According to Weill Cornell, liquid biopsy technology, which is based on blood tests for early cancer detection, has faced significant hurdles due to the challenges of identifying cancer’s mutational signatures from low concentrations of tumour DNA.
The Landau laboratory, situated at the Meyer Cancer Center of WCM and NYGC, claims to have dedicated nearly a decade to refining whole-genome sequencing methods to address these challenges, moving beyond targeted sequencing of expected mutation sites.
A 2024 study demonstrated the capability to identify advanced stages of melanoma and lung cancer through blood samples from patients, eliminating the need for sequencing data from tumour specimens.
US Tariffs are shifting - will you react or anticipate?
Don’t let policy changes catch you off guard. Stay proactive with real-time data and expert analysis.
By GlobalDataThe latest study leverages the cost-effectiveness of the new sequencing platform, and the researchers demonstrated the platform’s capability to identify tumour DNA at parts-per-million concentration levels, using known mutational patterns in patient tumours as a reference.
All samples were collected with informed consent from the subjects involved.
Researchers then improved the approach’s “accuracy” by including the error-correction method, which leverages redundant data in natural two-stranded DNA.
This combo method yielded extremely low error rates, suggesting the possibility of using the technique on blood samples without the requirement for tumour accessibility.
Partnerships with other research teams have shown the method’s potential in detecting and evaluating minimal cancer levels in individuals with bladder cancer and melanoma using only blood samples.
This research received support from the National Cancer Institute of the National Institutes of Health.
Further backing came from the Melanoma Research Alliance Established Investigator Award, the Mark Foundation Aspire Award, and the Burroughs Wellcome Fund Career Award for Medical Scientists.
NewYork-Presbyterian/Weill Cornell Medical Center urologic oncologist and Englander Institute for Precision Medicine chief research officer Dr Bishoy Faltas said: “This collaboration allowed us to analyse circulating tumour DNA from patients with bladder cancer and identify the distinct mutational signatures that my lab has extensively studied.”